Lately, there have been reports in some sensationalist media that state that a document delivered by Pfizer to the FDA and published by order of Judge Mark Pittman of the US Federal Court, which can be downloaded and read in its entirety here, is practically a confession by Pfizer, through the which is stating that its mRNA vaccines against COVID-19 contain graphene oxide, a highly toxic and conductive substance that has the ability to allow electricity to pass through its structure.
Graphene is conductive because it has this property due to the arrangement of carbon atoms in its structure, which forms a flat hexagonal lattice with very strong covalent bonds between them. Due to this structure, graphene is highly conductive of electricity and heat. This conductive property can be dangerous in certain contexts, especially if the graphene comes into contact with substances that can react with it and release toxic compounds.
What The Exposé claims in particular in a headline is the following: BREAKING: FDA confirms graphene oxide is in COVID-19 mRNA vaccines after being forced to release confidential Pfizer documents by US Federal Court order
Later in the article, a subheading reads: "But they were lying to you"
What is true in the news is the part where it says: The FDA had tried to delay the release of Pfizer's Covid-19 vaccine safety data for 75 years, despite approving the injection after only 108 days of a security review on December 11, 2020.
A group of scientists and medical researchers sued the FDA under FOIA, to force the release of hundreds of thousands of documents related to the license of the Pfizer-BioNTech Covid-19 vaccine.
Federal Judge Mark Pittman ordered the FDA to release 55,000 pages per month, and PHMPT has since posted all documents on its website as they are released.
What is inaccurate is that one of the most recent documents released by the FDA, saved as 125742_S1_M4_4.2.1 vr vtr 10741.pdf, confirms that page 7 of the document delivered to the Federal Court says that reduced graphene oxide is required to manufacture the Pfizer Covid-19 vaccine.
But... What does page 7 of this document really say?
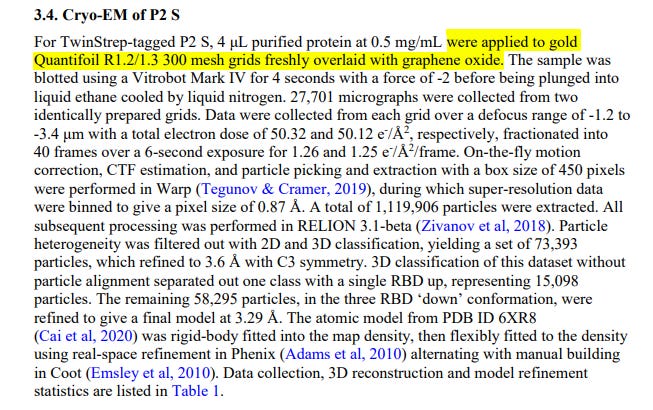
What the part of the document that is the subject of controversy says is recognized in point 3.4. Cryo-EM which refers to the method Pfizer used to study the protein. It literally says: “P2 S Cryo-MS For TwinStrep labeled P2 S, 4 μL of 0.5 mg/mL purified protein was applied to freshly overlaid 300 mesh Quantifoil R1.2/1.3 gold grids with graphene oxide.”
What does the above mean?
P2S Cryo-EM is the cryoelectron microscopy technique used to image the spike glycoprotein known as 2spike” (P2S). Cryoelectron microscopy is a microscopy technique used to study biological structures at the molecular level.
TwinStrep Labeling is a type of labeling used to purify Spike protein. A special amino acid sequence is introduced into the protein, which contains a TwinStrep tag which is a tool used in biology to purify proteins and study their function. The TwinStrep tag is made up of two parts, one part that binds to a protein called Strep-Tactin and one part that binds to another protein called Strep-Tag. The Strep-Tactin protein binds to the TwinStrep tag very specifically, allowing the TwinStrep tagged protein to be purified.
TwinStrep is like a magnet with two parts, one that attracts a protein called Strep-Tactin and the other that attracts another protein called Strep-Tag. The protein to be studied is bound to the part of the tag that attracts the Strep-Tag, and then the TwinStrep-tagged protein is purified using the part of the tag that attracts the Strep-Tactin.
The sample is passed through the solid matrix, and the Spike protein is attached to the Strep-Tactin through the TwinStrep tag.
Unwanted impurities are removed by washing the solid matrix.
The purified protein is eluted from the solid matrix. When purifying a protein, a solid matrix is generally used to trap the protein in this case the Spike and separate it from other components of the sample. This is accomplished by adding a special solution that changes the conditions of the solid matrix, causing the protein to detach and collect in a new sample. The entire elution process is the method of releasing the purified protein from the solid matrix in which it has been bound during the purification process.
The advantage of the TwinStrep tag is that it binds to the protein in a very specific and stable way, allowing the protein to be studied with great precision and separated from other proteins that may be present in the sample.
The Pfizer paper says they used 4 µL of 0.5 mg/mL purified protein applied to 300 mesh Quantifoil R1.2/1.3 gold grids freshly overlaid with graphene oxide. This indicates the specific conditions used to prepare the samples for study in the electron microscope.
In this particular study, what Pfizer is saying is that they used Quantifoil R1.2/1.3 gold grids freshly overlaid with graphene oxide, which means that they used this graphene as a support for the protein sample. As graphene is a very thin and resistant material, it is a good support in the preparation of samples for cryo-EM. As graphene is an excellent electrical conductor, it allows a better dissipation of the electrical charge during the microscopy process, which improves the quality of the images obtained and allows the protein structure to be seen in high-quality images.
In the study of proteins and biomolecules, graphene is used as a support to hold and stabilize the sample during observation under a cryoelectron microscope.
So for a better understanding, imagine an optical microscope, the crystals where the products to be observed are supported are the supports where what is to be observed is held. In this way it is easier to understand that when gold Quantifoil R1.2/1.3 grids of 300 mesh are used, just superimposed with graphene oxide, they are explaining to you what material the support is made of where the protein has been attached to observe it in this cryomicroscopy electronics.
In the case of the Spike glycoprotein study, graphene oxide-coated graphene was used to anchor and stabilize the TwinStrep-tagged purified protein. The sample is dried and snap frozen before being viewed under a cryoelectron microscope. This support is used for its ability to reduce radiation and electron absorption, its high thermal and electrical conductivity, and its ability to form bonds with a wide range of biological molecules, which facilitates the observation of proteins and biomolecules at the atomic level.
Graphene is a two-dimensional crystalline substance made up of carbon atoms arranged in a hexagonal structure. It is the thinnest material known, it is very resistant and a good conductor.
To understand how these proteins work, they have studied their structure in detail. This is achieved using techniques such as X-ray crystallography, which allows high-resolution images of proteins to be obtained.
The protein crystallization process is very complex and requires a support to keep the proteins stable. This is where the graphene discussed in the Pfizer paper came into play, since graphene is an excellent support for protein crystallization due to its chemical stability, low roughness, and ability to hold proteins in place without interfering. with its structure they decided to use it as a support for Spike to better understand how this protein works. Therefore, what this document says is that Pfizer has used graphene as a support to develop a support for electron microscopy of proteins and biomolecules.
Having explained what was the object of error in its interpretation, the text of point 3.4 of the document says: Cryo-MS of P2 S For P2 S labeled with TwinStrep, 4 μL of purified protein at 0.5 mg/mL were applied to grids gold Quantifoil R1.2/1.3 mesh 300 freshly overlaid with graphene oxide. The sample was dried using a Vitrobot Mark IV for 4 seconds at -2 force before immersing it in liquid ethane cooled with liquid nitrogen. 27,701 micrographs were collected from two identically prepared grids. Data was collected from each grid in a blur range of -1.2 to 090177e195e446ea\Approved\Approved on: 27-Dec-2020 02:23 (GMT)- 3.4 μm with a total electron dose of 50.32 and 50.12 e-/A2, respectively, divided into 40 frames in a 6-second exposure for 1.26 and 1.25 e-/A2/frame. On-the-fly motion correction, CTF estimation, and particle selection and extraction with a 450-pixel box size were performed in Warp (Tegunov & Cramer, 2019), during which super-resolution data was pooled to give a pixel size of 0.87 Å. A total of 1,119,906 particles were extracted. All post processing was done in RELION 3.1-beta (Zivanov and others, 2018). Particle heterogeneity was filtered with 2D and 3D classification, yielding a pool of 73,393 particles, which were refined to 3.6 Å with C3 symmetry. 3D classification of this dataset without particle alignment separated a class with a single RBD above, representing 15,098 particles. The remaining 58,295 particles, in the 'down' conformation of the three RBDs, were refined to give a final model at 3.29 Å. The atomic model of PDB ID 6XR8.
In other words, scientists took a very small sample of a protein we call Spike and put it on a very thin piece of a thing called a "grid" (graphene). Then, they dried the sample with a special machine and immersed it in a cold liquid.
The sentence that follows after the description of the process of applying the sample to grids with graphene oxide is "The sample was dried using a Vitrobot Mark IV for 4 seconds at a force of -2 before immersing it in liquid ethane cooled with liquid nitrogen. ". This is the only mention of graphene in the entire document and it is not stated anywhere else in the document, or at any time that the vaccine contains graphene in this brief that Pfizer delivered to the Federal Court.
27,701 micrographs were collected from two identically prepared grids, and data was collected from each grid.
The data was collected in a blur range of -1.2 to 3.4 µm with a total electron dose of 50.32 and 50.12 e-/A2, respectively, the lack of sharpness in the image means that a range was used. of specific defocusing to obtain the images. On-the-fly motion correction, CTF estimation, and particle selection and extraction with a box size of 450 pixels in Warp were performed. These are the processes used to process the images obtained during cryoelectron microscopy.
A total of 1,119,906 particles were extracted, number of particles extracted from the images for further analysis. Post-processing was performed in RELION 3.1-beta, which is a state-of-the-art electron microscopy image processing software. In this step, the heterogeneity of the particles was filtered by 2D and 3D classification to obtain a set of 73,393 particles, which were refined to 3.6 Å with C3 symmetry. This is the number of particles that were refined and the resolution obtained in Ångstroms (unit of measurement used to measure the resolution of images obtained by cryoelectron microscopy.
3D classification of this dataset without particle alignment separated a class with a single RBD above, representing 15,098 particles. The remaining 58,295 particles, in the 'down' conformation of the three RBDs, were refined to give a final model at 3.29 Å. RBD stands for "receptor binding domain," and refers to the part of the protein that binds to the host cell's receptor. In this case, the atomic model can be found in PDB ID 6XR8. The PDB is an online database containing information on the three-dimensional structure of proteins and other macromolecules. The structures are resolved experimentally using techniques such as X-ray crystallography or cryoelectron microscopy. Each structure is assigned a unique identifier known as a PDB ID. When it is said that the atomic model of a protein can be found in PDB ID 6XR8, it means that the three-dimensional structure of the protein has been determined by a structure-solving technique such as X-ray crystallography or cryoelectron microscopy and that this structure has been registered in the PDB database with the unique identifier 6XR8. The atomic model is a detailed representation of the protein showing the position of each atom in the structure. Information that can be used to understand how the protein works and how it interacts with other molecules in the body.
For all these reasons, the statements that circulate in some sensationalist media are false and have been misinterpreted.
Pfizer states on page 7 of the study in section 3.4
https://expose-news.com/2023/04/02/fda-confirms-graphene-is-in-the-covid-vaccines/
https://icandecide.org/wp-content/uploads/2023/02/125742_S1_M4_4.2.1-vr-vtr-10741.pdf




Yes. Thank you.! Expose knew this was false and refused to retract. Why should they.! They were shilling for funds at the top of their articles. Sensationalism is very detrimental to truth.